Metabolomics
Mass spectrometry (MS)-based metabolomics
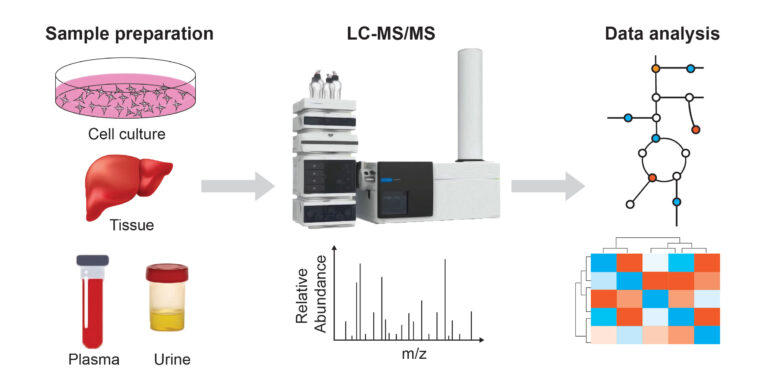
Mass spectrometry-based metabolomics has revolutionized our understanding of cellular metabolism in the past couple of decades. We use liquid chromatography-mass spectrometry (LCMS)-based metabolomics to identify and quantify metabolites from biological samples, including cells, biofluids, and tissues. Our platform uses an Agilent 6546 QTOF high-resolution mass spectrometer coupled with an Agilent 1290 infinity II UHPLC. We are currently optimizing methods to identify ~500 metabolites using hydrophilic interaction liquid chromatography (HILIC) and reverse-phase liquid chromatography (RPLC). We perform stable isotope tracing experiments using labeled metabolites (13C glucose, 13C glutamine, etc.) to gain insights into the fates of metabolites in normal and perturbed cellular states.
We are integrating our strong expertise in organic chemistry and enzymology with LCMS to develop novel metabolomics methods to identify otherwise difficult-to-identify unknown metabolites in untargeted metabolomics.
The Patgiri lab metabolomics platform is actively seeking suitable collaborations that require targeted metabolomics and/or stable isotope tracing experiments. Please email Anupam (anupam.patgiri@emory.edu) for details.
Publications that used our metabolomics platform
1. Zlatic, S.A., Werner, A., Surapaneni, V., Lee, C. E., Gokhale, A., Singleton, K., Duong, D., Crocker, A., Gentile, A., Middleton, F., Dalloul, J. M., Liu, W., Patgiri, A., Tarquinio, D., Carpenter, R., Faundez, V. “Systemic proteome phenotypes reveal defective metabolic flexibility in Mecp2 mutants” Human Molecular Genetics, (2023) (Patgiri lab co-authors underlined)